Palindromes in the genome

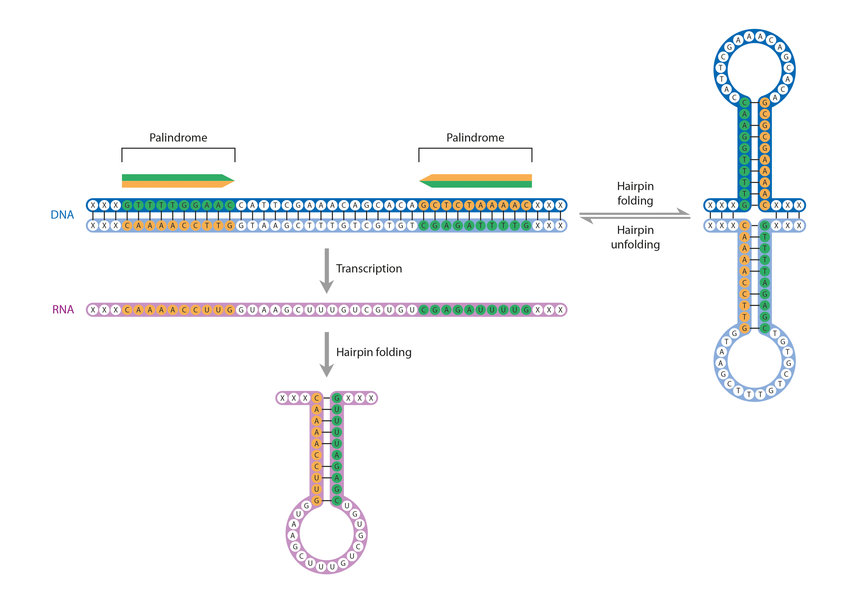
“Able was I ere I saw Elba”: this rather bombastic statement is a palindrome sentence, in other words it reads exactly the same forwards as it does backwards. The beginning of the CRISPR revolution was marked by the discovery of a large number of repeated palindromic sequences in a region of bacterial DNA. In these sequences, the letters of the genetic code, the four base molecules adenine, cytosine, thymine and guanine, are ordered such that they have the same order as the second complementary DNA-strand – in this case read in opposite direction. This is the property that gives CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) its tongue-twisting name.
Unlike word palindromes like ‘civic’ and ‘tenet’, which have a meaning, the palindromes in the genetics dictionary do not make sense and cannot be translated into functional proteins. Nevertheless, they are not entirely meaningless. DNA-cutting proteins frequently use palindrome sequences as recognition sequences, at which they cut the DNA molecule. These sequences can be four, six or eight base pairs long, although some cutting proteins require 20 or more base pairs.
Out of the palindromic sequences of the CRISPR region RNA molecules are transcribed, which adopt a very stable arrangement (secondary structure). They range between 23 and 47 base pairs in length. Variable regions of similar length can be found between these sequences. They originated from the genome of foreign DNA that penetrated the bacterial cell, and are also known as spacer DNA.
The CRISPR region includes a promoter which ensures that the CRISPR region can be read and translated into the CRISPR-RNA (crRNA). Other genes known as CRISPR-associated genes (Cas) are located adjacent to it. These genes provide the blueprint for the Cas proteins - namely, the enzymes that cut the DNA strand. The CRISPR and spacer sequences are followed by a region for an RNA molecule known as tracrRNA, which guides the cutting molecules and the crRNA to their target locations on the virus DNA.