Functioning of CRISPR-Cas9

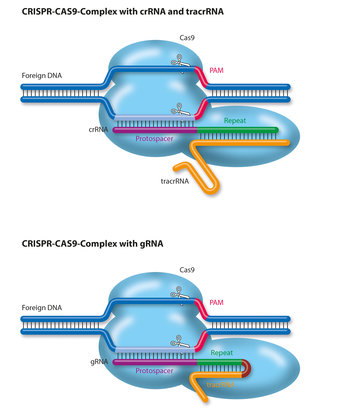
With the CRISPR-Cas9 system, the CRISPR- and spacer sequences are translated into the CRISPR-RNA (crRNA). Before the latter can lead the Cas9 protein to the cutting site on the DNA, it must be converted into its final form by cutting enzymes and parts must be removed from it. RNase III is such an enzyme. Together with the tracrRNA sequence it transforms the original form of the crRNA into a mature and functional molecule.
The finished crRNA contains a transcript of the CRISPR sequence and the foreign DNA. This provides Cas9 with the recognition sequence at which the cutting molecule should cleave the DNA. The crRNA binds with the tracrRNA, as only the two together can show Cas9 the way to the cutting site.
The recognition sequence that matches the crRNA is not sufficient on its own for Cas9 to be able to bind to DNA: it also needs a ‘proto-spacer adjacent motif’ or PAM. Cas9 can only bind to the DNA strand if a three-letter sequence consisting of two guanines and any base is located adjacent to the recognition sequence.
The two strands of DNA uncoil and the crRNA/tracrRNA molecule – or an artificial guide RNA – can attach itself. The enzyme then cuts the two DNA strands in the same place. Consequently, to be able to cut DNA, Cas9 needs both the recognition sequence and a PAM.