Further development of CRISPR-Cas9
Scientists all over the world are working intensely on gaining a better understanding of how CRISPR-Cas9 works and developing the gene scissors for use in research and medical practice. They are also continuing the search for other molecular tools that may work even more effectively than CRISPR-Cas9.
Greater accuracy

Despite the accuracy with which the gene scissors locates its target site on the DNA, it does sometimes miss. For example, the shorter the recognition sequence, the greater the probability that such a sequence will arise several times in the genome and Cas will cut the DNA in several locations. In addition, CRISPR-Cas9 sometimes makes mistakes and docks at other sites in addition to its target sequence.
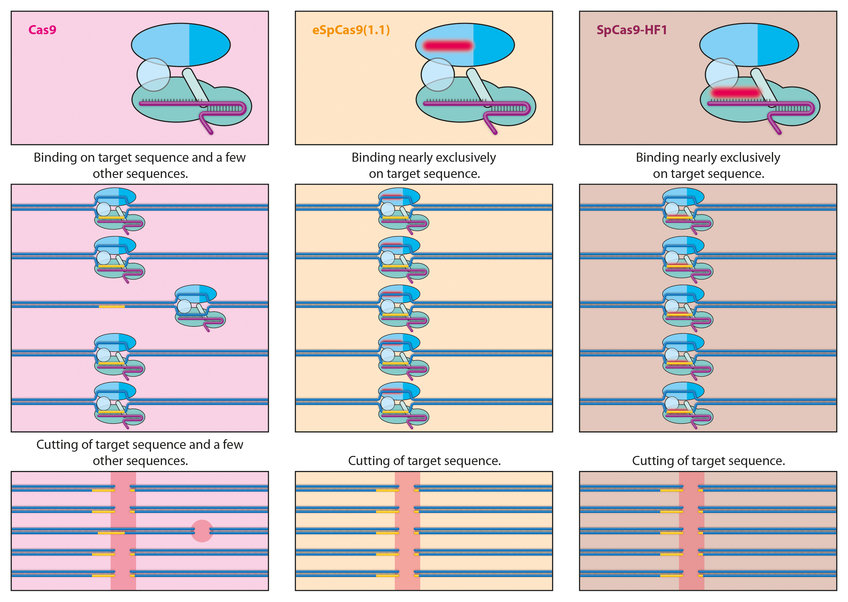
For this reason, researchers would like to modify the system to ensure that such misses become even rarer. To this end, they are using new amino acids in the Cas9 protein and changing its architecture as a result. For example, using the eSpCas9 and Cas9-HF1 variants, scientists have developed extremely precise Cas9 proteins which, in the case of HF1, achieve an accuracy rate of over 99.9 percent. Further tests will be needed to show whether this is sufficient. When it comes to modifying the DNA of millions or even billions of cells in the human body, an error rate as low as 0.01 percent would still have considerable impacts.
Another attempt to increase the accuracy of the gene scissors involves proteins called nickases, which only cleave one of the two twisted DNA strands. To make a complete cut in the DNA, two enzymes with their guide RNAs must therefore bind at the same DNA sequence. This corresponds to a kind of ‘dual control principle’ and should reduce the risk of errors in the cutting of the DNA.
Researchers may be able to replace individual letters of the genetic code in future using a programmable CRISPR-Cas9 system. To do this, they combined CRISPR-Cas9 with a cytidine deaminase, which is an enzyme that can transform cytidine components in the DNA into uridine. This results in the letter C of the genetic alphabet (cytosine) being transformed into a T (thymine), and a G (guanine) being transformed into an A (adenine).
Led by a guide RNA, the new CRISPR-Cas9 variant can carry out a base exchange and in this way correct point mutations. At least 3,000 serious diseases arise through such point mutations in which a T is replaced by a C, or an A by a G. The scientists tested their development on a variant of the apoE gene, which increases the risk of developing Alzheimer’s and only differs from the normal form of the gene by a single letter. They succeeded in providing 75 percent of the cells with the Alzheimer’s variant with the healthy form of the gene. However, they are not yet able to define the letters to be exchanged with extreme precision – only within sequences that are five bases in length.
Scissors for RNA
Scientists not only want to improve the existing gene editing system, they would also like to find alternatives that work even more accurately and are easier to manage. Cpf1 is an example of such an alternative. This enzyme is also part of the bacterial immune system and is used to cut foreign DNA. Tests have shown that Cpf1 can also cut RNA. In the bacterial cells, Cpf1 starts by removing individual sequences of the crRNA molecule and thereby acts as a mature protein. This eliminates the need for additional proteins like RNase III. The mature crRNA then guides Cpf1 to its target sequence on the DNA.
Cpf1 thus has a dual function: first, it makes the crRNA capable of functioning; second, it cuts the DNA at the region recognized by the crRNA. Moreover, unlike Cas9, Cpf1 is not reliant on the help of a tracrRNA to reach its target site. The structure of this system is therefore even simpler than that of CRISPR-Cas9. Moreover, unlike Cas9, Cpf1 does not cut the two DNA strands at exactly the same place. This makes it easier for researchers to add new sequences at these ‘overhanging’ sequences.
Many diseases are the result of excessively high or insufficient volumes of proteins. Such defects can be treated by modifying RNA molecules. RNA molecules can also be more suitable targets in cases where the repair mechanisms of cells are so strong that they prevent the DNA from being edited. Unlike changes to the DNA, modifications to the RNA are not permanent and are not usually inherited. Thus, RNA editing does not bring about changes in organisms that are passed down through the generations. This poses fewer ethical risks as a result.
In addition to Cpf1, researchers have discovered another enzyme, C2c2, that can cut the RNA molecule accurately. The bacterium Leptotrichia shahii uses C2c2 to render viruses with RNA as genetic material harmless.
Scientists can now even cut RNA using CRISPR-Cas9. To do this they add a short DNA molecule with a PAM to the guide RNA. The two bind with each other so that the guide RNA can no longer dock at DNA and only at RNA. PAMs are usually short sequences on the target DNA, which CRISPR-Cas9 needs to dock at its actual recognition sequence. In this way, a DNA scissors becomes a cutting tool for RNA.
Whether the further development of CRISPR-Cas9 will play an important role in gene editing still remains to be seen. In any event, the search for new gene editing tools is in full swing.